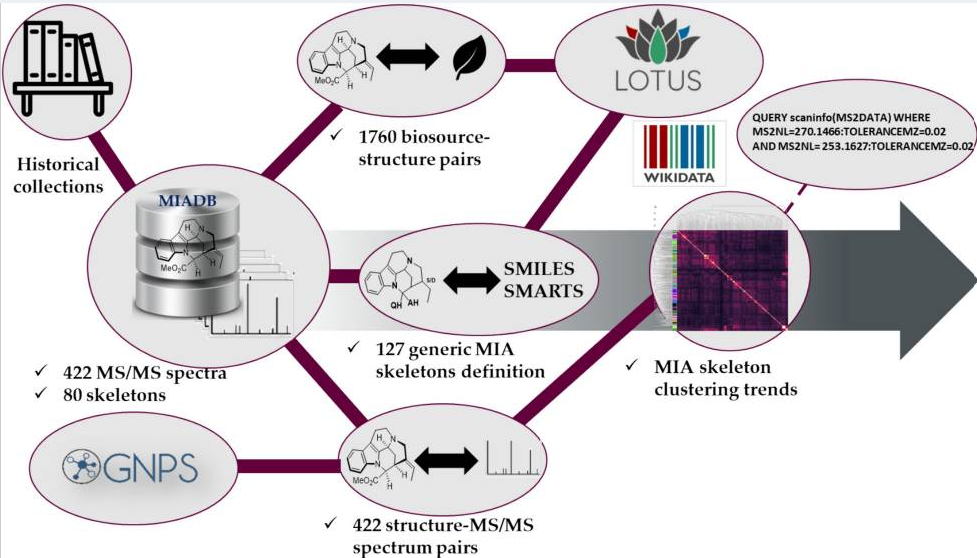
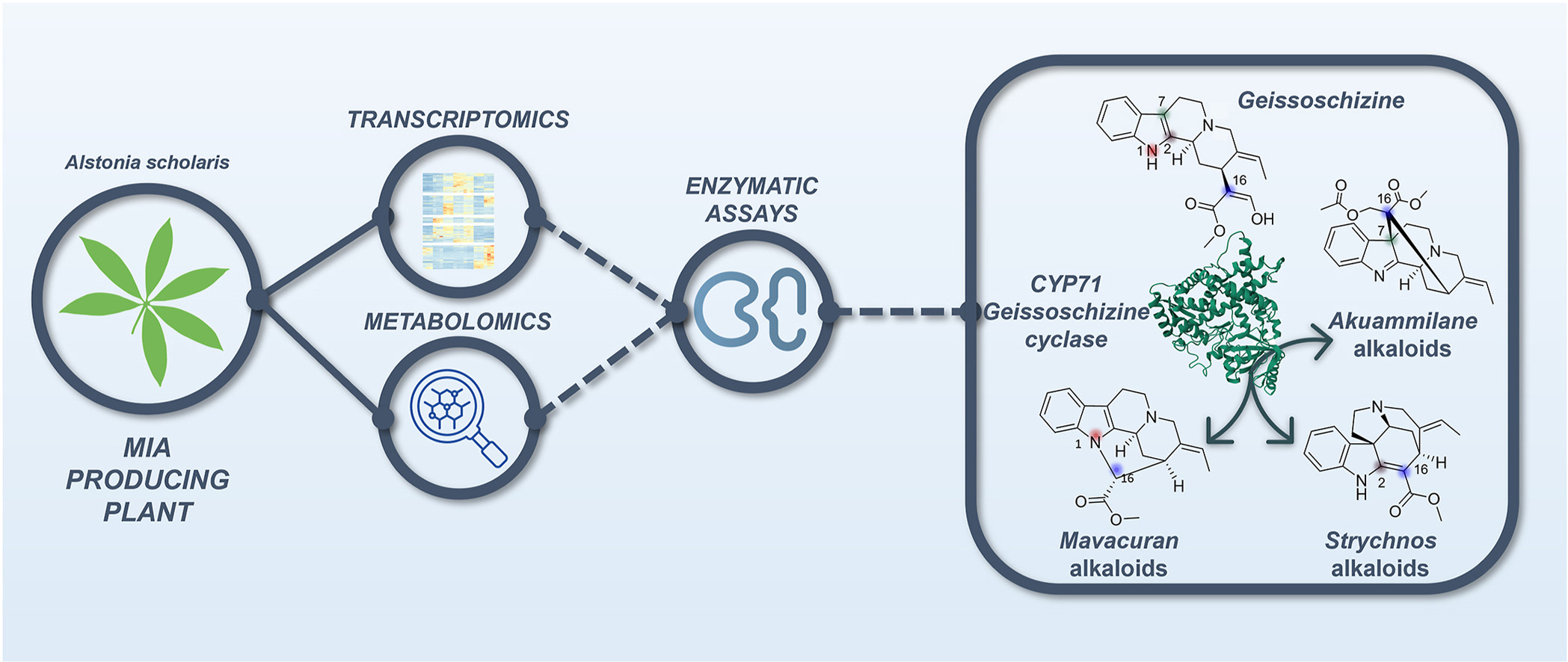
Vinca minor, also known as the lesser periwinkle, is a well-known species from the Apocynaceae, native to central and southern Europe. This plant synthesizes monoterpene indole alkaloids (MIAs), which are a class of specialized metabolites displaying a wide range of bioactive- and pharmacologically important properties. Within the almost 50 MIAs it produces, V. minor mainly accumulates vincamine, which is commercially used as a nootropic. Using a combination of Oxford Nanopore Technologies long read- and Illumina short-read sequencing, a 679,098 Mb V. minor genome was assembled into 296 scaffolds with an N50 scaffold length of 6 Mb, and encoding 29,624 genes. These genes were functionally annotated and used in a comparative genomic analysis to establish gene families and to investigate gene family expansion and contraction across the phylogenetic tree. Furthermore, homology-based MIA gene predictions together with a metabolic analysis across four different V. minor tissue types guided the identification of candidate MIA genes. These candidates were finally used to identify MIA gene clusters, which combined with synteny analysis allowed for the discovery of a functionally validated vincadifformine-16-hydroxylase, reinforcing the potential of this dataset for MIA gene discovery. It is expected that access to these resources will facilitate the elucidation of unknown MIA biosynthetic routes with the potential of transferring these pathways to heterologous expression systems for large-scale MIA production.